Shiny Application to Iowa DNR MSIM-SGCN Modeling
Xiaodan (Annie) Lyu
Join work with Tyler M. Harms
September 23, 2016
Introduction
Period: Oct 2015 - May 2016
State Wildlife Grants - Iowa Department of Natural Resources
Run predictive models using R package RMark
Produce predictive maps using ArcGIS
Develop an interactive web application using Shiny
Outline
Project overview: data source and objectives
Methods: model fit and covariates
Model Validation: AUC
Results: example of estimates table and predictive maps
Demonstrate Shiny application
Data Collection
Multiple Species Inventory and Monitoring (MSIM) Program
- Iowa DNR and Iowa State University
Sampling Interval
Primary: 2006-2014
Secondary: survey occasions (days)
Survey Objects
414 SGCN, only 69 SGCN where sufficient data were available
Birds, mammals, reptiles, amphibians, odonates and butterflies
Objectives
Predict the distribution of species of conservation need
- robust design occupancy model
Create predictive species maps for priority SGCN
ArcGIS raster files
Shiny interactive web application
Prioritize areas of conservation action for SGCN
- habitat restoration and management
Methods
Robust design occupancy model (MacKenzie et al. 2003)
- package RMark
Parameters of Interest
probability of occupancy ( ψψ )
probability of colonization ( γγ )
probability of extinction ( ϵϵ )
probability of detection ( pp )
Model types
- RDOccupEG, RDOccupPE, RDOccupPG, ...
Statistical Model
Pr(Xi)=ϕ0{T=1∏t=1D(pX,t)ϕt}pX,TPr(Xi)=ϕ0{T=1∏t=1D(pX,t)ϕt}pX,T
- pX,tpX,t : vector denoting probability of observing the detection history Xi,tXi,t in primary period tt
- ϕtϕt : matrix of transition probabilities between states of occupancy from primary period tt to t+1t+1 ϕt=[P(Xt+1=1|Xt=1)P(Xt+1=0|Xt=1)P(Xt+1=1|Xt=0)P(Xt+1=0|Xt=0)]=[1−ϵtϵtγt1−γt],t=1,...,T-1ϕt=[P(Xt+1=1|Xt=1)P(Xt+1=0|Xt=1)P(Xt+1=1|Xt=0)P(Xt+1=0|Xt=0)]=[1−ϵtϵtγt1−γt],t=1,...,T-1
ϕ0=[ψ11−ψ1]ϕ0=[ψ11−ψ1]
Example
Pr(Xi,1=010)=ψ1(1−p1,1)p1,2(1−p1,3)Pr(Xi,2=000|Xi,1)=(1−ϵ1)3∏j=1(1−p2,j)+ϵ1Pr(Xi=010 000)=ϕ0D(p010,1)ϕ1p000,2=[ψ11−ψ1][(1−p1,1)p1,2(1−p1,3)000] ×[1−ϵ1ϵ1γ11−γ1][∏3j=1(1−p2,j)1]=ψ1(1−p1,1)p1,2(1−p1,3)[(1−ϵ1)3∏j=1(1−p2,j)+ϵ1]Pr(Xi,1=010)=ψ1(1−p1,1)p1,2(1−p1,3)Pr(Xi,2=000|Xi,1)=(1−ϵ1)3∏j=1(1−p2,j)+ϵ1Pr(Xi=010 000)=ϕ0D(p010,1)ϕ1p000,2=[ψ11−ψ1][(1−p1,1)p1,2(1−p1,3)000] ×[1−ϵ1ϵ1γ11−γ1][∏3j=1(1−p2,j)1]=ψ1(1−p1,1)p1,2(1−p1,3)[(1−ϵ1)3∏j=1(1−p2,j)+ϵ1]
Statistical Model (cont'd)
Including Covariates θ=exp(Z′β)1+exp(Z′β)
θ is the probability of interest
Z is the matrix of covariate information
β is the vector of logistic model coefficients to be estimated
Covariates
Landscape habitat variables [1]
Radius of sampled site: 200m, 500m and 1km
Land use classification:
Water, Wetland, Grassland, Woodland and AgricultureLandscape configuration:
percentage of landscape (PLAND), large patch index (LPI), edge density (ED), patch density (PD) and interspersion-juxtaposition (IJI)
Climate variables [2]
- Wind speed (km/h), Cloud cover (%), Temperature ( oC )
[1] site-specific, modeled on ψ,γ and ϵ
[2] time-varying, modeled on p
Model Selection and Validation
Akaike's Information Criterion adjusted for small sizes ( AICc )
- Models with ΔAICc≤2⇒ strong support
(Burnham and Anderson 2002)
- Models with ΔAICc≤2⇒ strong support
Area under the receiver operating characteristic curve (AUC)
evaluate performance of predicting occupancy
training data set: survey years 2006-2012, 2014
test data set: survey year 2013 (better representative)
package pROC
AUC=0.5⇒ random guess
AUC=1.0⇒ perfect prediction
(Jimenez-Valverde 2012)
ROC Curve
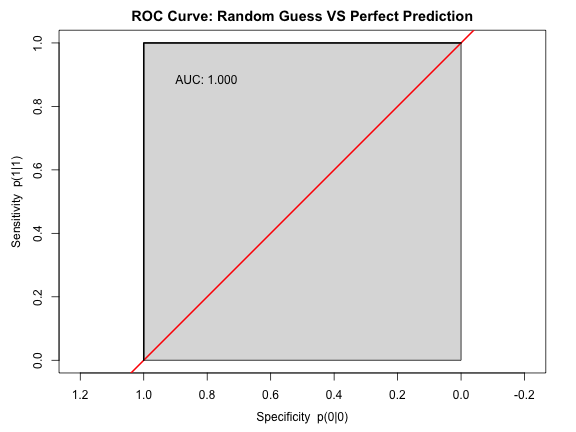
Results
Results Summary
| Statistics | Value/Range |
|---|---|
| Number of SGCN modeled | 64 out of 69 |
| Occupancy Prob ˆψ | 0.001(0.0003)~0.995(0.004) |
| Colonization Prob ˆγ | 0.0003(0.0001)~0.999(0.00007) |
| Detection Prob ˆp | 0.030(0.028)~0.998(0.006) |
| AUC values | 0.426~0.921 |
| Number of AUC>0.5 | 61 out of 64 |
Results (Cont'd)
Best Models for Each Specie
| Species | Model |
|---|---|
| Red-shouldered Hawk | ψ(Wod1KPLND) γ(Ag1KPD)p(Cld) |
| Yellow-billed Cuckoo | ψ(Wod500PLND) γ(Wod1kLPI)p(Wind) |
| Red-headed Woodpecker | ψ(Wod500PLND) γ(Ag500PD)p(Cld) |
| Eastern Wood-pewee | ψ(Wod500PLND) γ(Wod1KPLND)p(Wind) |
| Acadian Flycatcher | ψ(Wod500PLND) γ(Wod500PLND)p(Wind) |
| Veery | ψ(Wod1kED) γ(Ag500LPI)p(Wind) |
Results (Cont'd)
AUC and Coefficients Estimates under Best Model
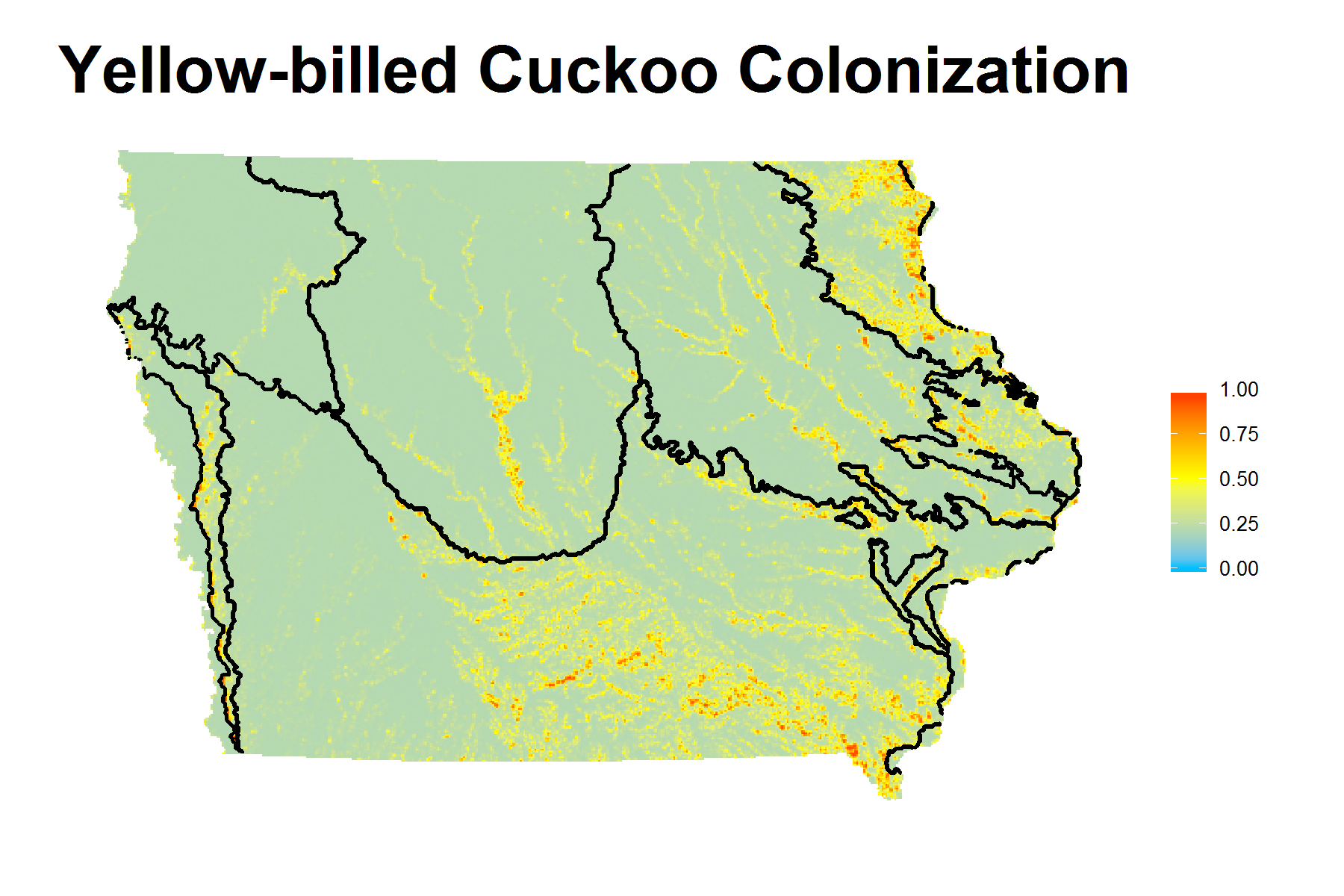
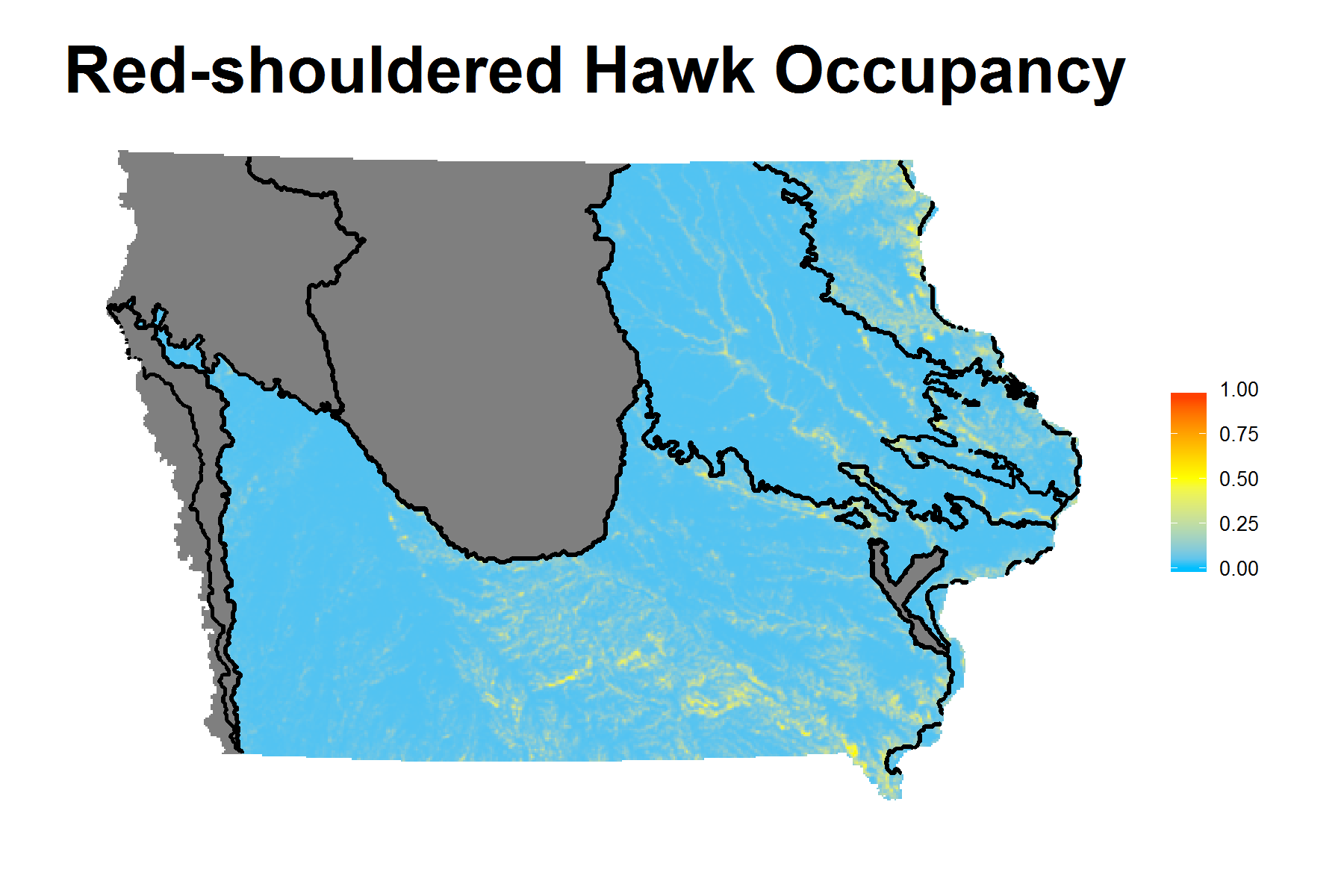
Shiny
Shiny by RStudio is a web application framework for R.
No HTML, CSS or JavaScript knowledge required to turn your analyses into interactive web applications.
install.packages("shiny")library("shiny")runExample("01_hello")ui.R defines the page layout and user interface
server.R contains the R code to create any output
More information available at Shiny Webpage
About Our Shiny Application
Interactive web application to display predictive maps and parameter estimates for each SGCN
Easy to download personalized data and maps
Available to researchers and managers across Iowa (credentials needed)
Hosted by CSSM for 1-2 years
The URL for accessing the application is https://dnrswg.cssm.iastate.edu/